Research Experience
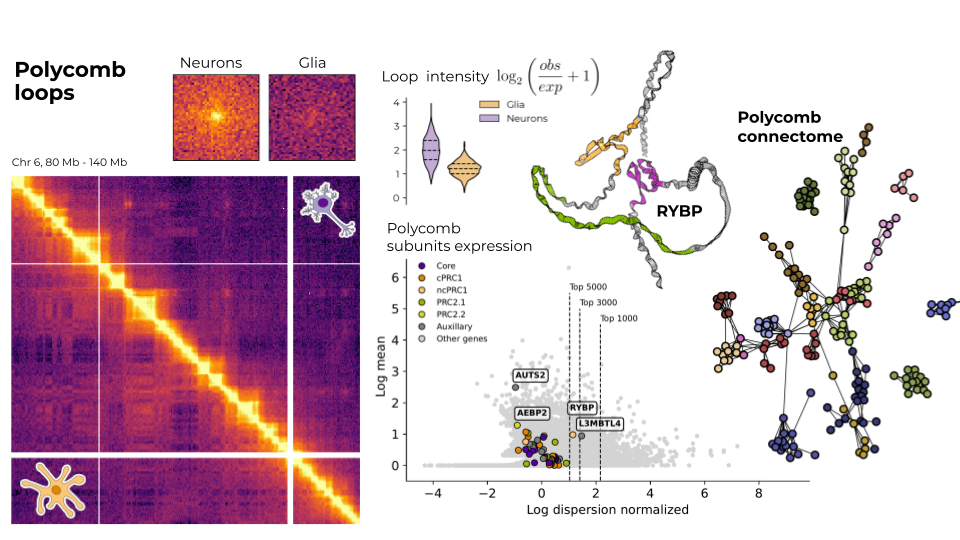
Research on human brain cells chromatin structure
I analyse different omics datasets (mainly Hi-C data, single-cell Hi-C and also ChIP-seq, ATAC-seq, RNA-seq) to study the differences in chromatin structure between neurons and glia. As my master's project, I am focusing on those features that are related to polycomb group proteins. However, I am globally interested in the chromatin and epigenetic changes landscape underlying the processes of neurogenesis and the development of brain disorders
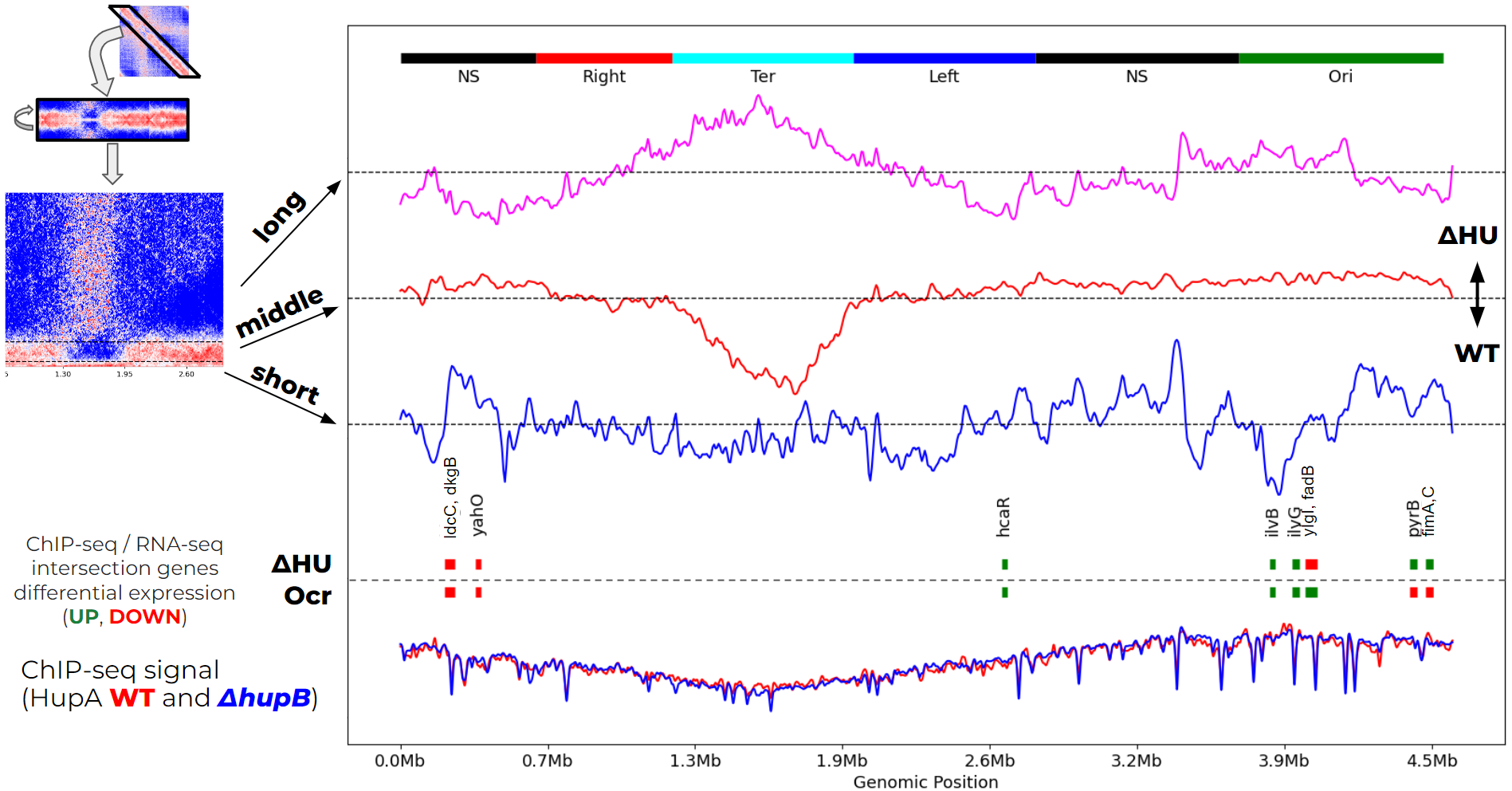
Phage DNA-mimic Ocr impact on the E. coli chromatin
I analyse Hi-C data of the E. coli genome and integrate it with the ChIP-seq and RNA-seq data. We found that Ocr DNA-mimic expression is not equal to the Hu E. coli DNA-structuring protein deletion (which actually made things more complicated).
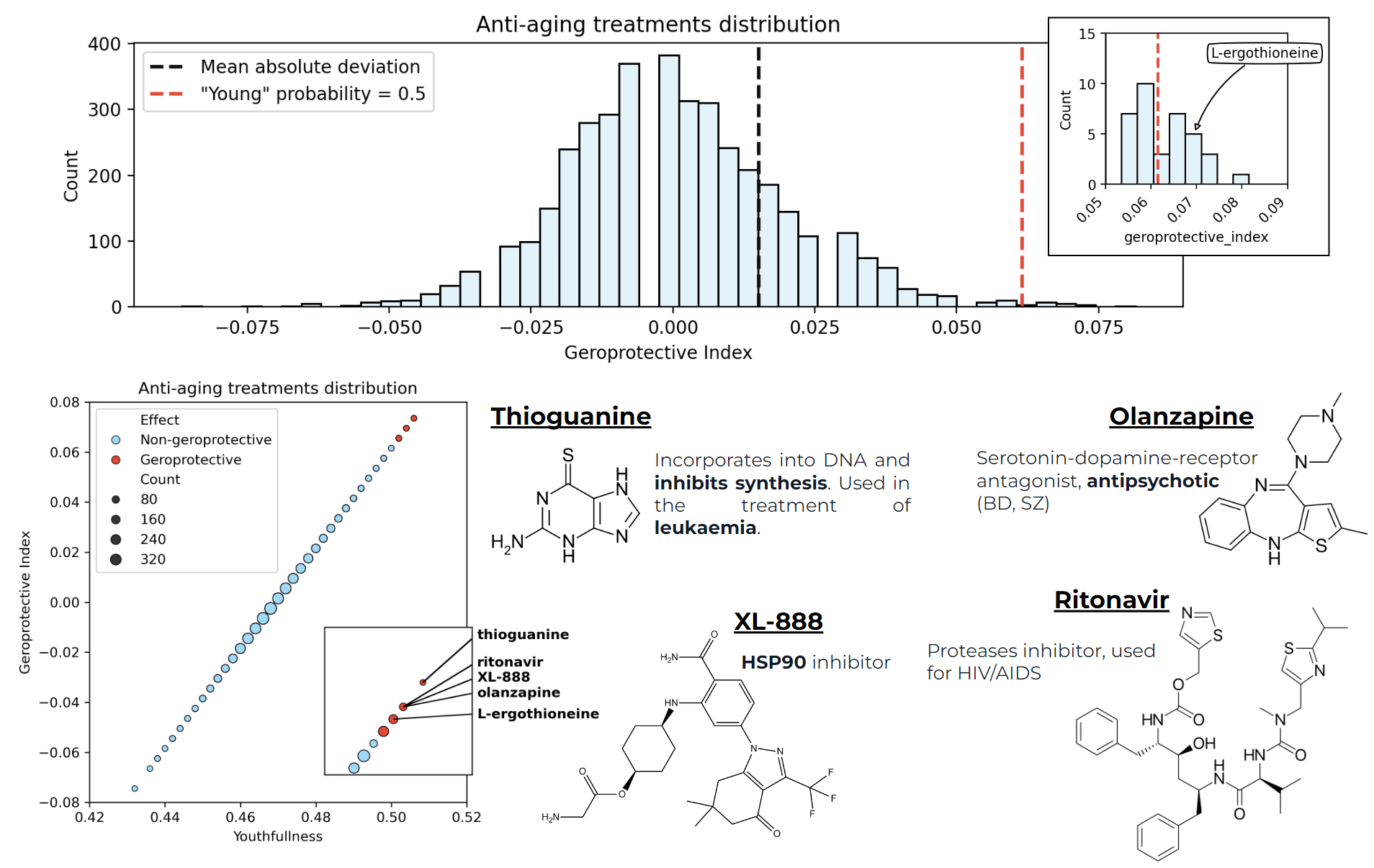
Transcriptomics-based longevity drug discovery
There was built a method for selecting potential anti-aging molecules based on a random forest model and two RNA-seq datasets (age-dependent and treatment-dependent transcriptome ) < (you can find out more here)
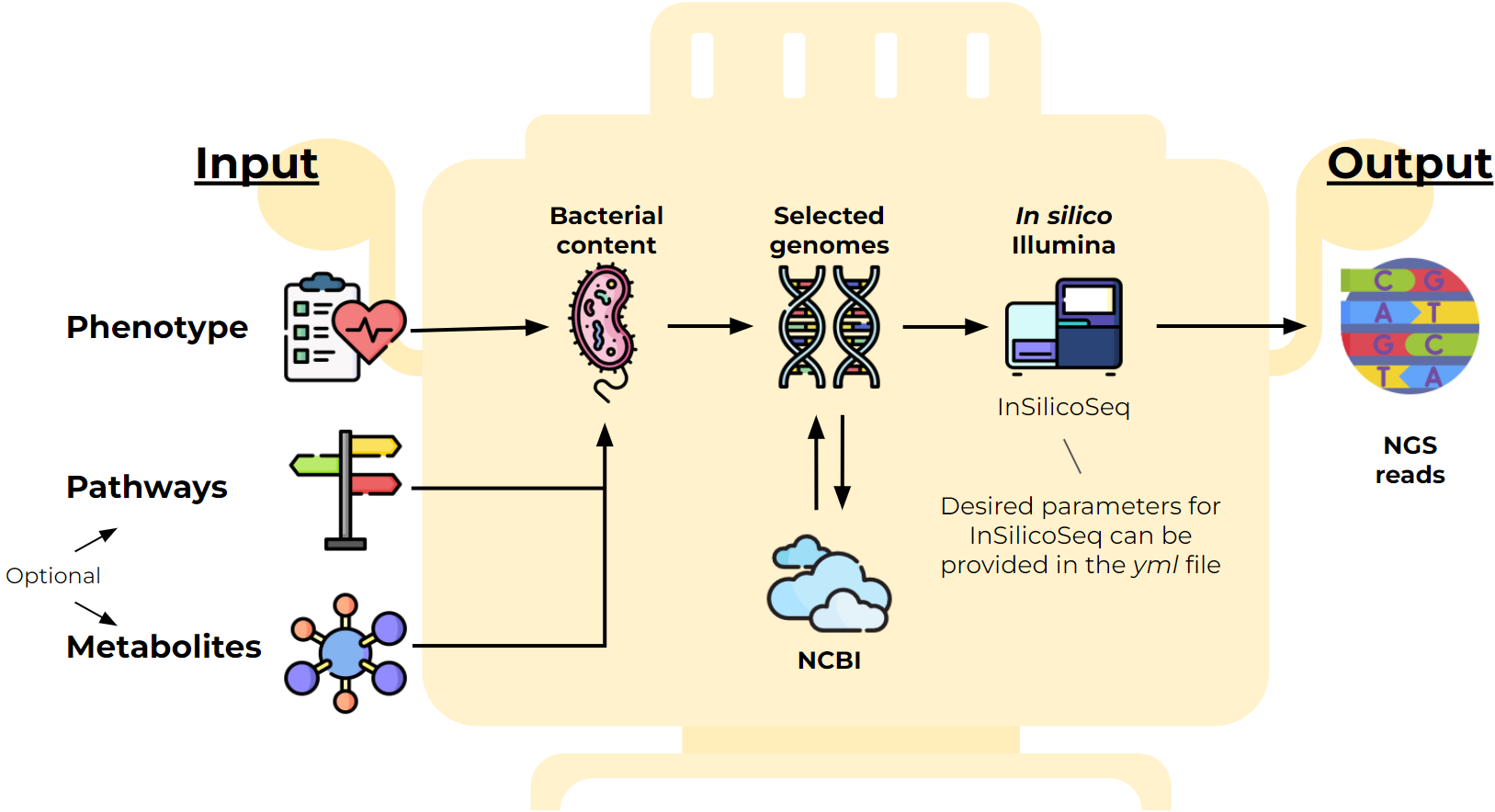
In silico generation of metagenomic communities
I develop a python tool (GitHub SAMOVAR) to generate metagenomes with specified properties. It is useful for validating the work of metagenome analysis tools. I implemented part of the algorithm for selecting the required species based on SAT-solvers as well as interaction with the NCBI database
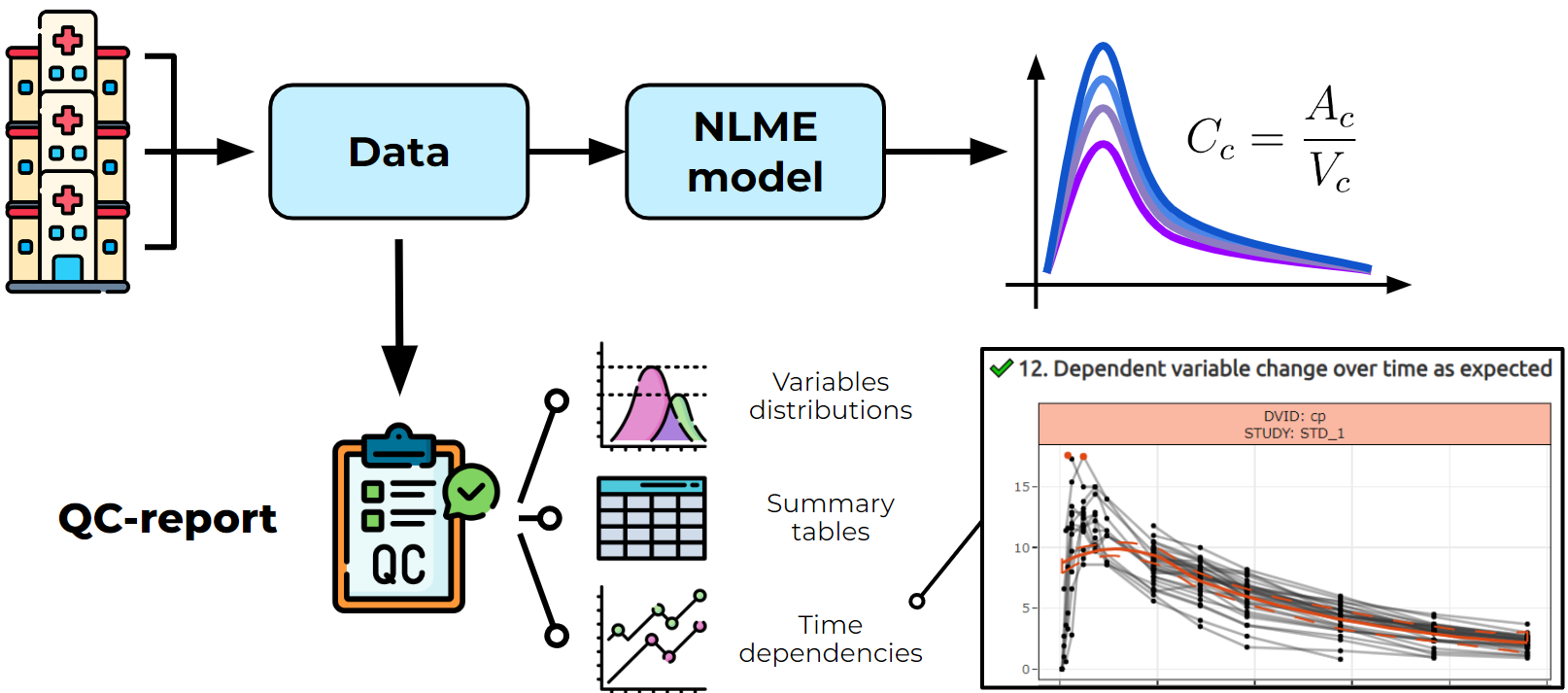
PHANDORIN: data quality check tool for non-linear mixed effects modeling
I developed an R-based tool PHANDORIN (PHArmacometrics Nlme Datasets Observation, Review and INspection ) for pharmacometrics data quality control (example report)
Research on type I CRISPR-Cas systems
I perform all stages of the work from conducting experiments and Illumina libraries preparation to NGS data processing. My key research topics are:
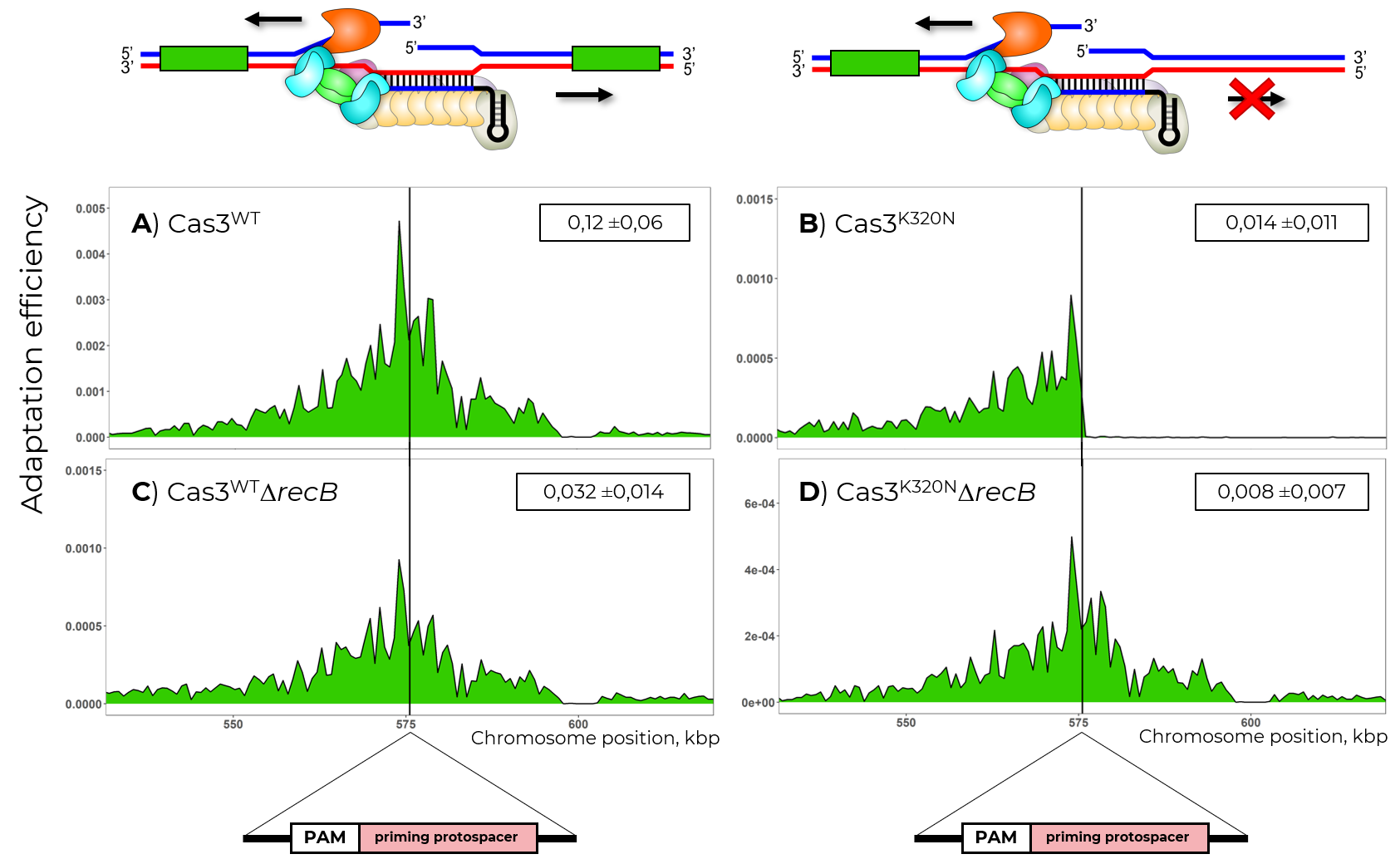
- Study of functioning of the Cas3 helicase-nuclease
- Studying the role of cellular nucleases in the operation of CRISPR-Cas systems
Institute of Molecular Genetics, National Research Centre «Kurchatov Institute»