Hi, i am
Nikita Vaulin
Bioinformatician. Biophysicist. Molecular biologist.

Bioinformatician. Biophysicist. Molecular biologist.

I am an omics-fan bioinformatician interested in cell fate decisions, especially in the context of neurodevelopment.
I think (I hope) we are now on the cusp of putting together a complete picture of how the various processes together govern the life of a cell.
Transcription factors, chromatin structure, post-translational modifications, etc.
- they set the rules of the game in which the cell must go from the opening through the mittelspiel to the endgame.
Chaos plays its role, but I believe that by defining a fully-omics state space we can anticipate all possible outcomes.
You can find me struggling with this questions in the Adameyko lab and Khrameeva lab. And also in Bioinformatics institute, where I teach python programming.
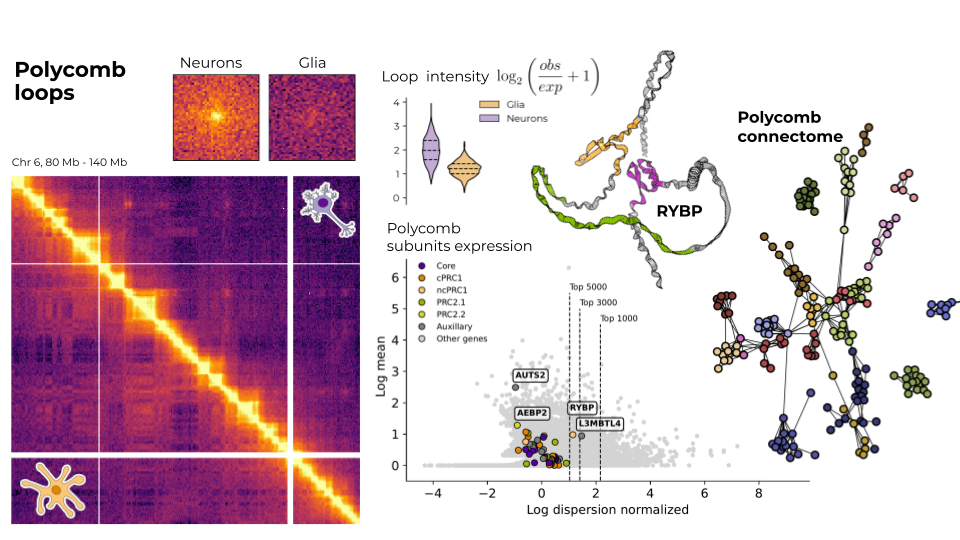
I analyse different omics datasets (mainly Hi-C data, single-cell Hi-C and also ChIP-seq, ATAC-seq, RNA-seq) to study the differences in chromatin structure between neurons and glia. As my master's project, I am focusing on those features that are related to polycomb group proteins. However, I am globally interested in the chromatin and epigenetic changes landscape underlying the processes of neurogenesis and the development of brain disorders
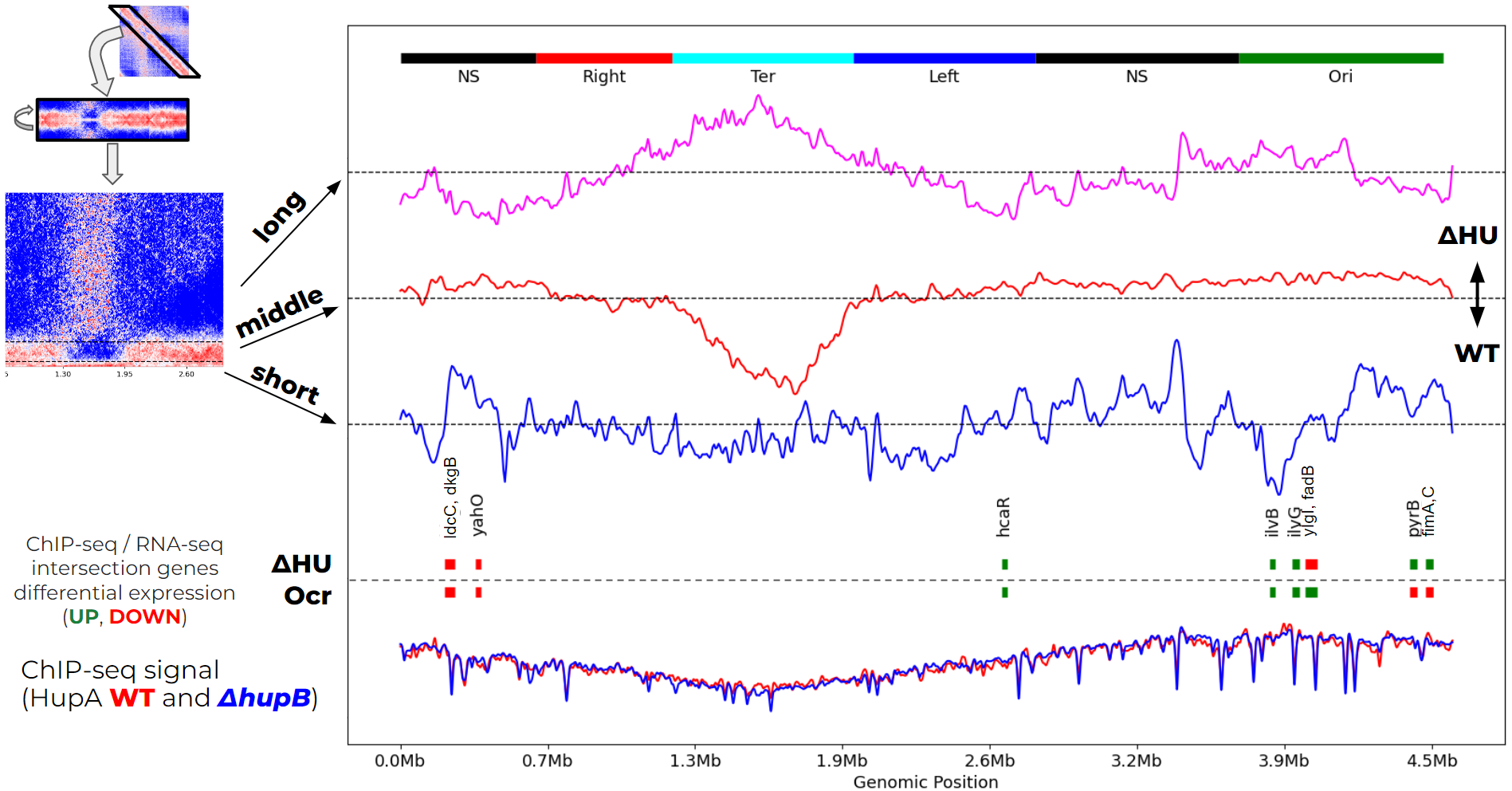
I analyse Hi-C data of the E. coli genome and integrate it with the ChIP-seq and RNA-seq data. We found that Ocr DNA-mimic expression is not equal to the Hu E. coli DNA-structuring protein deletion (which actually made things more complicated).
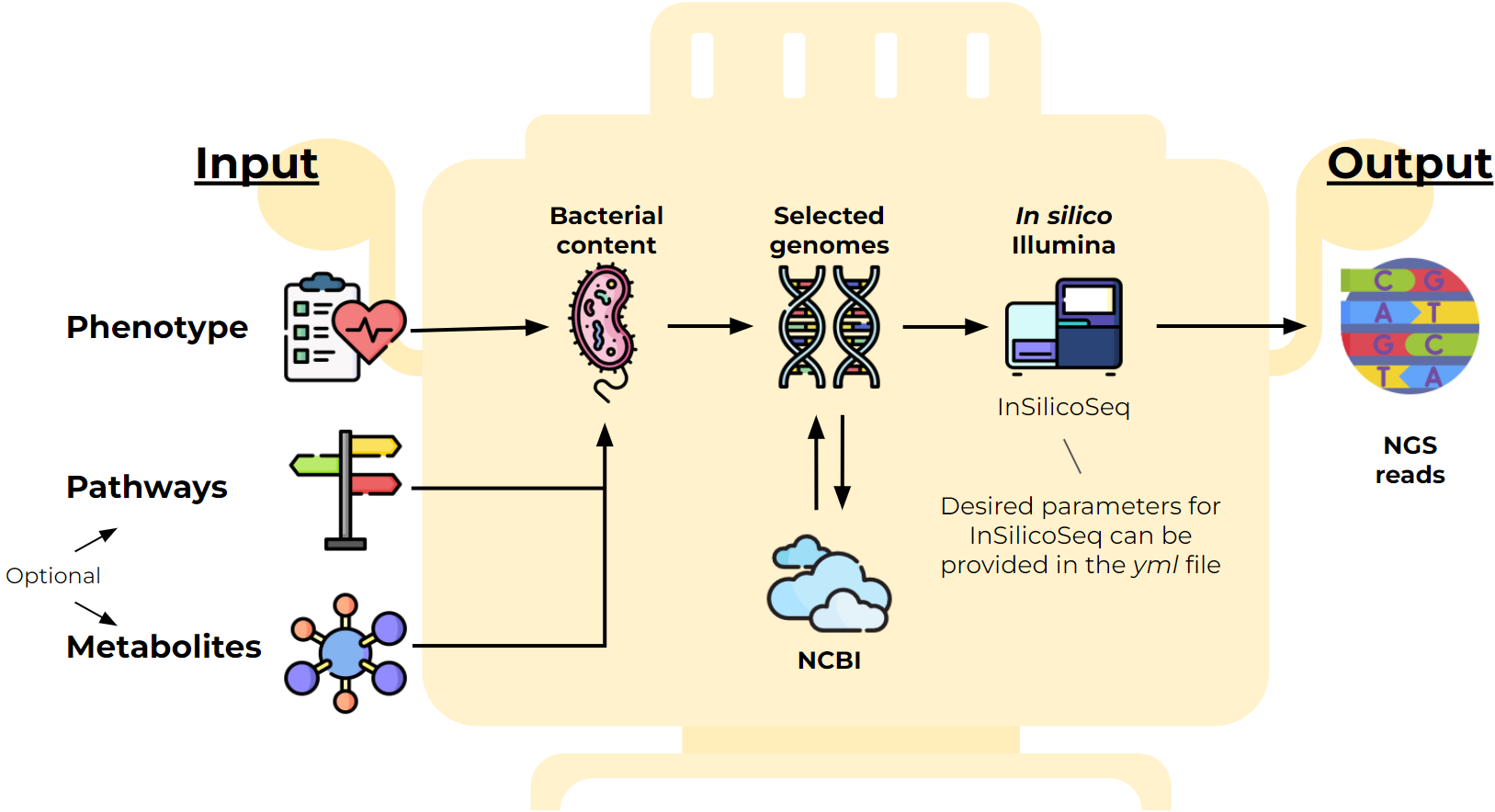
I develop a python tool (GitHub SAMOVAR) to generate metagenomes with specified properties. It is useful for validating the work of metagenome analysis tools. I implemented part of the algorithm for selecting the required species based on SAT-solvers as well as interaction with the NCBI database

Python (Pandas, NumPy, Scikit-learn) R (ggplot2, tidyverse, knitr, emo) Bash (scripts), Git Classical ML, basic C++
NGS data processing ChIP-seq, Hi-C, GWAS, RNA-seq variant calling, genome assembly biostatistics and visualization
NGS libraries preparation ChIP-seq 3C gene knock-out work with E. coli and P1 phage
laser cutter
I developed a Skoltech ISP (indepenent study period) course for 16 hours. In this course we practiced with some tools and shared each other experice. This course were awarded as a best student ISP 2023 course!

I teach a year-long programming course. We start with the basics of python and then move on may interesting and advanced things which are widely used in computational biology.
I teach a Hi-C data analysis as a part of the NGS data analysis workshop in BI. Also I give public lectures on the 3D genomics (example in russian).
I managed club meetings, gave short lectures on some topics, demonstrate solutions to some problems from the Rosalind website
Teaching assistant: